Navigation
What is qPTMplants
We developed the qPTMplants, an integrative database of quantitative post-translational modifications in plants, which curated and hosted the published 23 types of PTMs including 429,821 sites on 123,551 proteins in 43 plant species from 293 literature. Moreover, we collected and organized 620,509 kinds of quantitative events for 136,700 sites on 55,361 proteins from 139 literature. The detailed information about the PTM events was organized and visualized in detail. In addition, the annotations including the sequence and structural characteristics were orchestrated.
How to use qPTMplants
Quick Search
In the home page of qPTMplants, users can specify one field or all the fields to perform a quick search with the keyword(s) in the field(s). Users can simply click example and submit buttons to perform an example search.
Browse
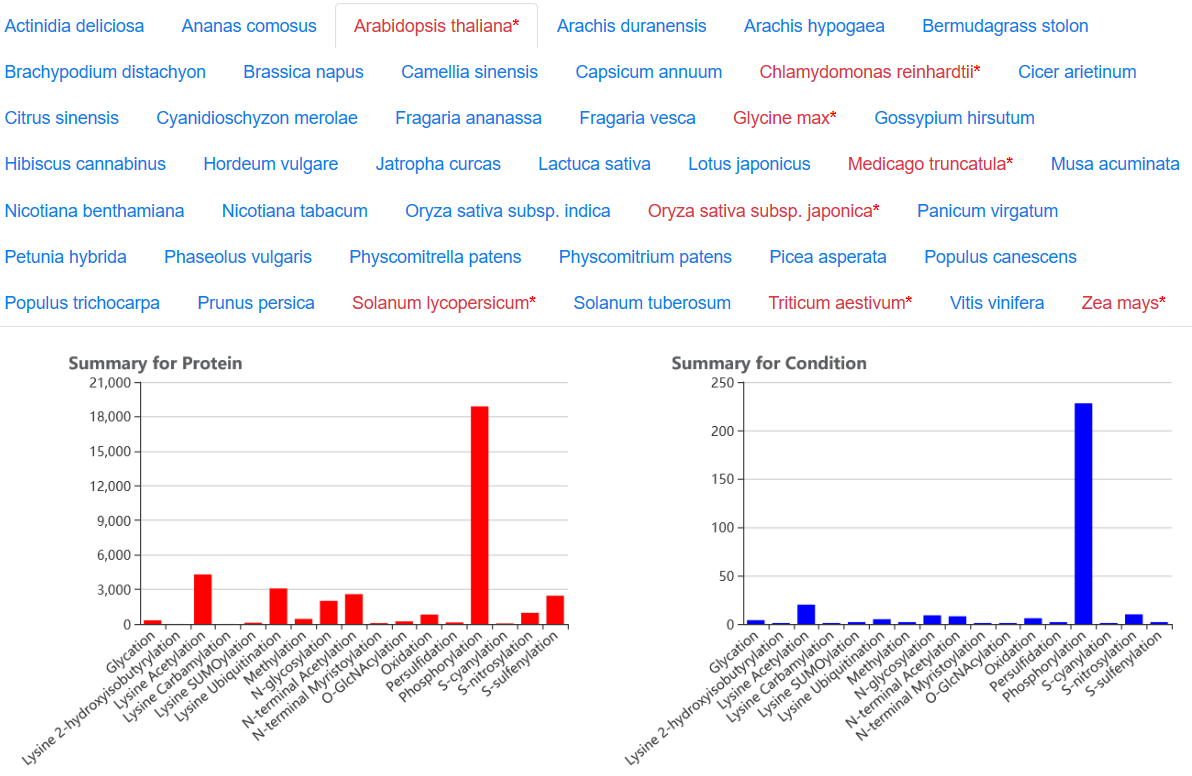
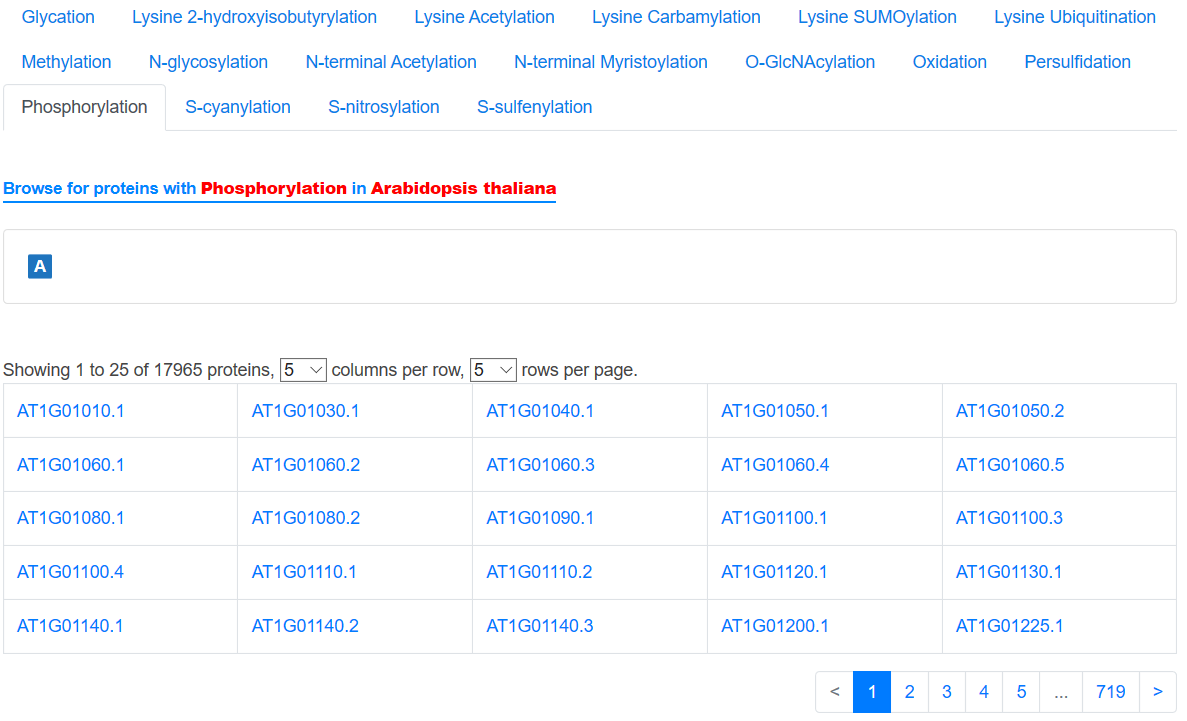
In the Browse page, users can click one term to search the specified data in qPTMplants. The data was firstly grouped by organisms and further grouped by PTMs, the detailed information is consisting of two modules including conditions and proteins, which are sorted alphabetically. Users can click the initial and select the term of interest.
Advanced Search
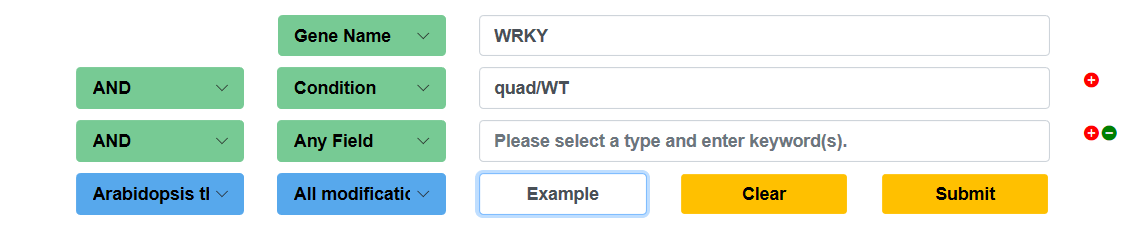
In the Search page, users can add more keyword boxes and choose 'AND', 'OR' or 'NOT' to perform a more accurate multiple keywords search. Users can simply click example and submit buttons to perform an example search.
The option of 'BLAST search' was designed to find related information in qPTMplants quickly. Users can input a protein sequence in FASTA format and select the required E value to search identical or homologous proteins through the blastall program in NCBI BLAST packages.

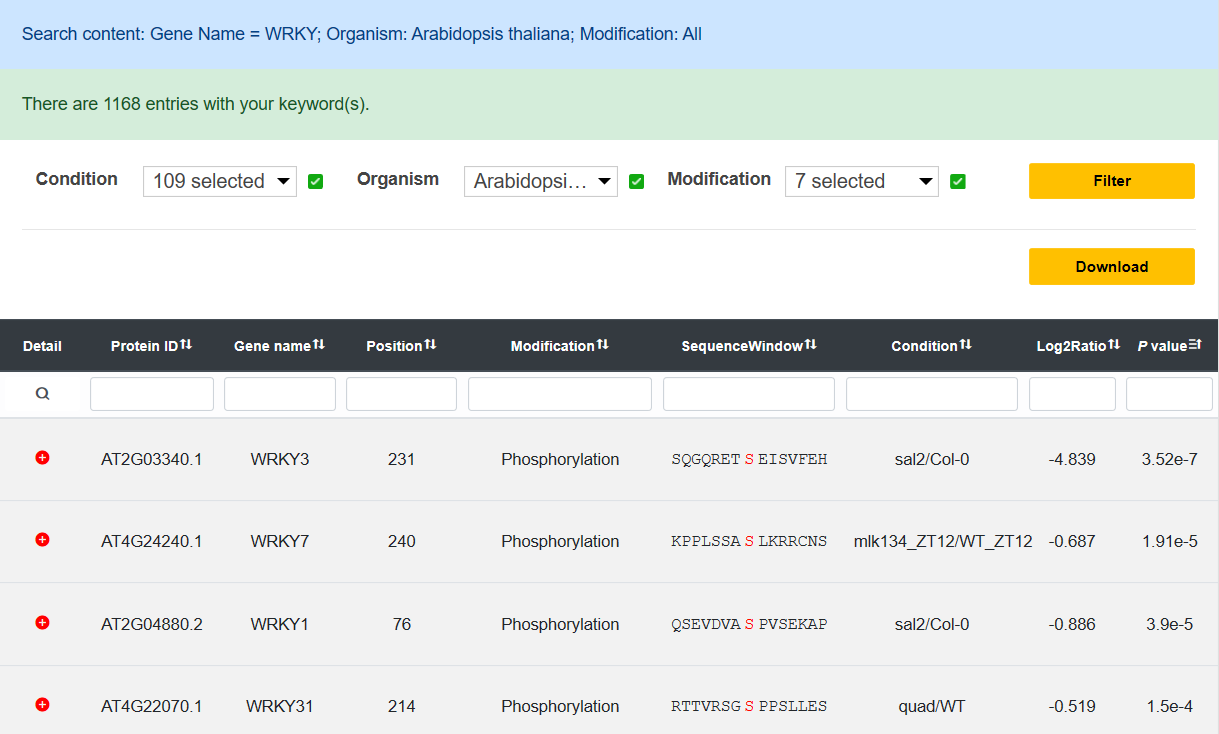
Result
In the Result page, users can view the search or browse results, and further filter them by the options we provide. More information about experiment and protein is also displayed while users click the 'Detail' button.
Post-translational modifications resources in plants
PhosPhAt
PhosPhAt is the Arabidopsis protein phosphorylation site database, which not only for experimental and predicted phosphorylation sites of individual proteins, but also for known substrates for a given kinase or kinase family.
website: http://phosphat.mpimp-golm.mpg.de
citation: Zulawski, M., Braginets, R. and Schulze, W.X. (2013) PhosPhAt goes kinases--searchable protein kinase target information in the plant phosphorylation site database PhosPhAt. Nucleic Acids Res., 41, D1176-1184. (PMID: 23172287)
P³DB
The plant protein phosphorylation database 3.0 (P3DB) has 47,923 phosphosites in 16,477 phosphoproteins curated across nine plant organisms from 32 studies, and incorporates multiple network viewers.
website: http://p3db.org
citation: Yao, Q., Ge, H., Wu, S., Zhang, N., Chen, W., Xu, C., Gao, J., Thelen, J.J. and Xu, D. (2014) P³DB 3.0: From plant phosphorylation sites to protein networks. Nucleic Acids Res., 42, D1206-1213. (PMID: 24243849)
Plant PTM Viewer
The Plant PTM Viewer is a central resource for exploring plant protein modifications, which comprises approximately 370,000 PTM sites for 19 types of protein modifications in plant proteins from five different species.
website: http://www.psb.ugent.be/PlantPTMViewer
citation: Willems, P., Horne, A., Van Parys, T., Goormachtig, S., De Smet, I., Botzki, A., Van Breusegem, F. and Gevaert, K. (2019) The Plant PTM Viewer, a central resource for exploring plant protein modifications. Plant J., 99, 752-762. (PMID: 31004550 )
FAT-PTM
The functional analysis tools for post-translational modifications (FAT-PTM) database includes eight different types of PTM and over 49,000 PTM sites identified in large-scale proteomic surveys of Arabidopsis thaliana. FAT-PTM currently supports tools to visualize protein-centric PTM networks, quantitative phosphorylation site data, PTM information displayed in protein-centric metabolic pathways and groups of proteins that are co-modified by multiple PTMs.
website: https://bioinformatics.cse.unr.edu/fat-ptm/
citation: Cruz, E.R., Nguyen, H., Nguyen, T. and Wallace, I.S. (2019) Functional analysis tools for post-translational modification: a post-translational modification database for analysis of proteins and metabolic pathways. Plant J., 99, 1003-1013. (PMID: 31034103)
dbPTM
dbPTM is a comprehensive database of experimental post-translational modifications, which has integrated over 900,000 PTM sites. With continuous updates, dbPTM also integrates annotation information such as the linear relationship between SAPs and PTM sites, the disease-associated PTM sites, and PTM crosstalk.
website: http://dbPTM.mbc.nctu.edu.tw/
citation: Huang, K.Y., Lee, T.Y., Kao, H.J., Ma, C.T., Lee, C.C., Lin, T.H., Chang, W.C. and Huang, H.D. (2019) dbPTM in 2019: exploring disease association and cross-talk of post-translational modifications. Nucleic Acids Res., 47, D298-d308. (PMID: 30418626)
SysPTM
SysPTM is a systematic resource installed with comprehensive PTM data including 471,109 PTM sites on 53,235 proteins, covering over 50 modification types across 2,031 species. Four web-based tools (PTMBlast, PTMPathway, PTMPhylog, PTMCluster) were implemented in SysPTM.
website: http://lifecenter.sgst.cn/SysPTM/
citation: Li, J., Jia, J., Li, H., Yu, J., Sun, H., He, Y., Lv, D., Yang, X., Glocker, M.O., Ma, L. et al. (2014) SysPTM 2.0: an updated systematic resource for post-translational modification. Database (Oxford), 2014, bau025. (PMID: 24705204)
EPSD
The eukaryotic phosphorylation site database (EPSD) collects 1,616,804 experimentally identified p-sites in 209,326 phosphoproteins from 68 eukaryotic species. These data not only come from published literature, but also integrate other resources, such as dbPPT, dbPAF, PhosphoSitePlus, etc.
website: http://epsd.biocuckoo.cn/
citation: Lin, S., Wang, C., Zhou, J., Shi, Y., Ruan, C., Tu, Y., Yao, L., Peng, D. and Xue, Y. (2021) EPSD: a well-annotated data resource of protein phosphorylation sites in eukaryotes. Brief. Bioinform., 22, 298-307. (PMID: 32008039)
iCysMod
iCysMod is an integrative database for protein cysteine modifications in eukaryotes, which curated and hosted 108,030 PCM events for 85,747 experimentally identified sites on 31,483 proteins from 48 eukaryotes for 8 types of PCMs.
website: http://icysmod.omicsbio.info
citation: Wang, P., Zhang, Q., Li, S., Cheng, B., Xue, H., Wei, Z., Shao, T., Liu, Z.X., Cheng, H. and Wang, Z. (2021) iCysMod: an integrative database for protein cysteine modifications in eukaryotes. Brief. Bioinform., 10.1093/bib/bbaa400. (PMID: 33406221)
PLMD
Protein lysine modification database (PLMD) integrates 284,780 modification events in 53,501 proteins from 176 eukaryotes and prokaryotes for 20 types of PLMs, involving ubiquitination, acetylation, sumoylation, and so on.
website: http://plmd.biocuckoo.org/
citation: Xu, H., Zhou, J., Lin, S., Deng, W., Zhang, Y. and Xue, Y. (2017) PLMD: An updated data resource of protein lysine modifications. J. Genet. Genomics, 44, 243-250. (PMID: 28529077)
